Step-by-step guide on using theDuring this walkthrough, we will become familiar with the mainMSTL ModelwithStatsforecast.
StatsForecast class and some relevant methods such as
StatsForecast.plot, StatsForecast.forecast and
StatsForecast.cross_validation in other.
The text in this article is largely taken from: 1. Changquan Huang •
Alla Petukhina. Springer series (2022). Applied Time Series Analysis and
Forecasting with
Python. 2.
Ivan Svetunkov. Forecasting and Analytics with the Augmented Dynamic
Adaptive Model (ADAM) 3. James D.
Hamilton. Time Series Analysis Princeton University Press, Princeton,
New Jersey, 1st Edition,
1994.
4. Rob J. Hyndman and George Athanasopoulos (2018). “Forecasting
Principles and Practice (3rd ed)”.
Table of Contents
- Introduction
- IMAPA Model
- Loading libraries and data
- Explore data with the plot method
- Split the data into training and testing
- Implementation of IMAPA with StatsForecast
- Cross-validation
- Model evaluation
- References
Introduction
The MSTL model (Multiple Seasonal-Trend decomposition using LOESS) is a method used to decompose a time series into its seasonal, trend and residual components. This approach is based on the use of LOESS (Local Regression Smoothing) to estimate the components of the time series. The MSTL decomposition is an extension of the classic seasonal-trend decomposition method (also known as Holt-Winters decomposition), which is designed to handle situations where multiple seasonal patterns exist in the data. This can occur, for example, when a time series exhibits daily, weekly, and yearly patterns simultaneously. The MSTL decomposition process is performed in several stages:- Trend estimation: LOESS is used to estimate the trend component of the time series. LOESS is a non-parametric smoothing method that locally fits data and allows complex trend patterns to be captured.
- Estimation of seasonal components: Seasonal decomposition techniques are applied to identify and model the different seasonal patterns present in the data. This involves extracting and modeling seasonal components, such as daily, weekly, or yearly patterns.
- Estimation of the residuals: The residuals are calculated as the difference between the original time series and the sum of the estimates of trend and seasonal components. Residuals represent variation not explained by trend and seasonal patterns and may contain additional information or noise.
MSTL
An important objective in time series analysis is the decomposition of a series into a set of non-observable (latent) components that can be associated with different types of temporal variations. The idea of time series decomposition is very old and was used for the calculation of planetary orbits by seventeenth century astronomers. Persons was the first to state explicitly the assumptions of unobserved components. As Persons saw it, time series was composed of four types of fluctuations:- a long-term tendency or secular trend;
- cyclical movements superimposed upon the long-term trend. These cycles appear to reach their peaks during periods of industrial prosperity and their troughs during periods of depressions, their rise and fall constituting the business cycle;
- a seasonal movement within each year, the shape of which depends on the nature of the series;
- residual variations due to changes impacting individual variables or other major events, such as wars and national catastrophes affecting a number of variables.
LOESS (Local Regression Smoothing)
LOESS is a nonparametric smoothing method used to estimate a smooth function that locally fits the data. For each point in the time series, LOESS performs a weighted regression using nearest neighbors. The LOESS calculation involves the following steps:- For each point t in the time series, a nearest neighbor window is selected.
- Weights are assigned to neighbors based on their proximity to t, using a weighting function, such as the Gaussian kernel.
- A weighted regression is performed using the neighbors and their assigned weights.
- The fitted value for point t is obtained based on local regression.
- The process is repeated for all points in the time series, thus obtaining a smoothed estimate of the trend.
MSTL General Properties
The MSTL model (Multiple Seasonal-Trend decomposition using LOESS) has several properties that make it useful in time series analysis. Here is a list of some of its properties:- Decomposition of multiple seasonal components: The MSTL model is capable of handling time series that exhibit multiple seasonal patterns simultaneously. You can effectively identify and model different seasonal components present in the data.
- Flexibility in detecting complex trends: Thanks to the use of LOESS, the MSTL model can capture complex trend patterns in the data. This includes non-linear trends and abrupt changes in the time series.
- Adaptability to different seasonal frequencies: The MSTL model is capable of handling data with different seasonal frequencies, such as daily, weekly, monthly, or even yearly patterns. You can identify and model seasonal patterns of different cycle lengths. (see) Seasonal periods
| Frecuencia |
|---|
| Data | Minute | Hour | Day | Week | Year |
|---|---|---|---|---|---|
| Daily | 7 | 365.25 | |||
| Hourly | 24 | 168 | 8766 | ||
| Half-hourly | 48 | 336 | 17532 | ||
| Minutes | 60 | 1440 | 10080 | 525960 | |
| Seconds | 60 | 3600 | 86400 | 604800 | 31557600 |
- Ability to smooth noise and outliers: The smoothing process used in LOESS allows to reduce the impact of noise and outliers in the time series. This can improve detection of underlying patterns and make it easier to analyze trend and seasonality.
- Improved forecasting: By decomposing the time series into seasonal, trend, and residual components, the MSTL model can provide more accurate forecasts. Forecasts can be generated by extrapolating trend and seasonal patterns into the future, and adding the stochastic residuals.
- More detailed interpretation and analysis: The MSTL decomposition allows you to analyze and understand the different components of the time series in a more detailed way. This facilitates the identification of seasonal patterns, changes in trend, and the evaluation of residual variability.
- Efficient Implementation: Although the specific implementation may vary, the MSTL model can be calculated efficiently, especially when LOESS is used in combination with optimized calculation algorithms.
Loading libraries and data
Tip Statsforecast will be needed. To install, see instructions.Next, we import plotting libraries and configure the plotting style.
| Time | Ads | |
|---|---|---|
| 0 | 2017-09-13T00:00:00 | 80115 |
| 1 | 2017-09-13T01:00:00 | 79885 |
| 2 | 2017-09-13T02:00:00 | 89325 |
| 3 | 2017-09-13T03:00:00 | 101930 |
| 4 | 2017-09-13T04:00:00 | 121630 |
-
The
unique_id(string, int or category) represents an identifier for the series. -
The
ds(datestamp) column should be of a format expected by Pandas, ideally YYYY-MM-DD for a date or YYYY-MM-DD HH:MM:SS for a timestamp. -
The
y(numeric) represents the measurement we wish to forecast.
| ds | y | unique_id | |
|---|---|---|---|
| 0 | 2017-09-13T00:00:00 | 80115 | 1 |
| 1 | 2017-09-13T01:00:00 | 79885 | 1 |
| 2 | 2017-09-13T02:00:00 | 89325 | 1 |
| 3 | 2017-09-13T03:00:00 | 101930 | 1 |
| 4 | 2017-09-13T04:00:00 | 121630 | 1 |
(ds) is in an object format, we need
to convert to a date format
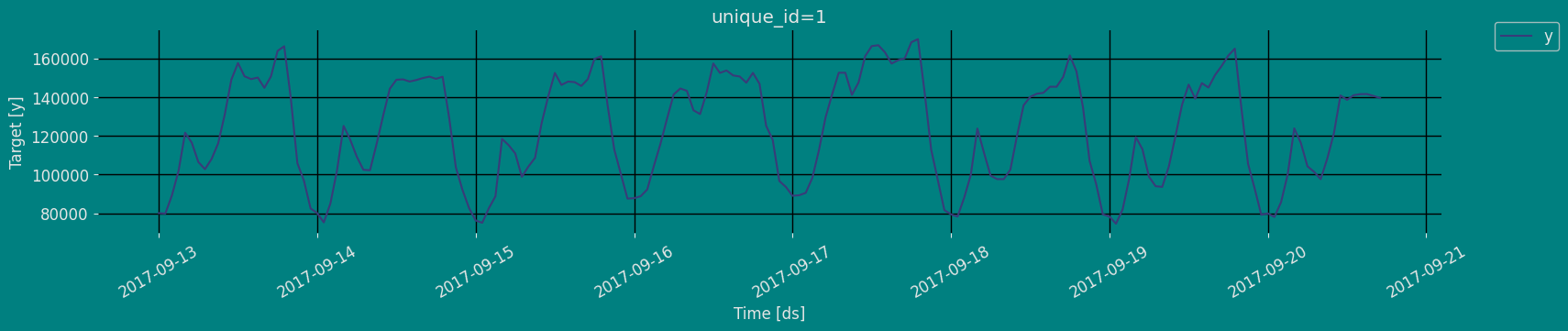
Explore Data with the plot method
Plot some series using the plot method from the StatsForecast class. This method prints a random series from the dataset and is useful for basic EDA.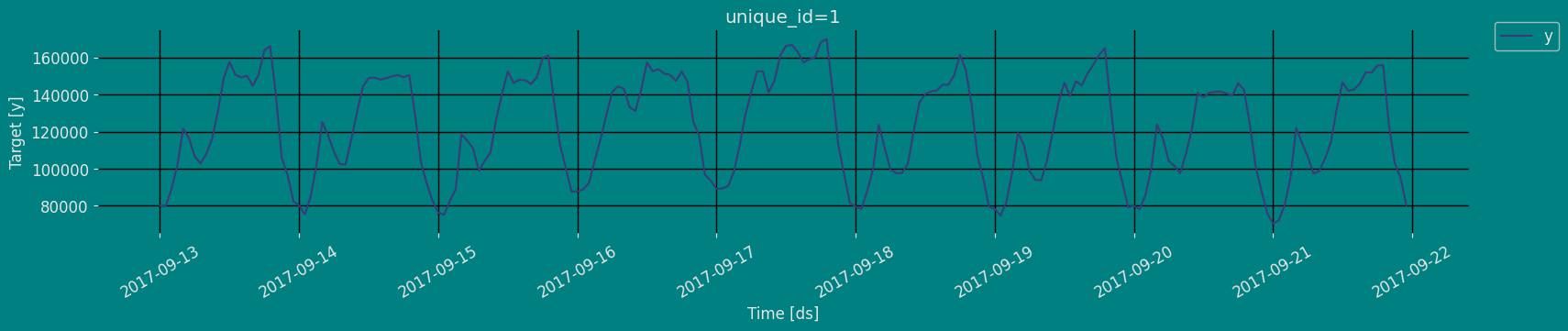
Autocorrelation plots
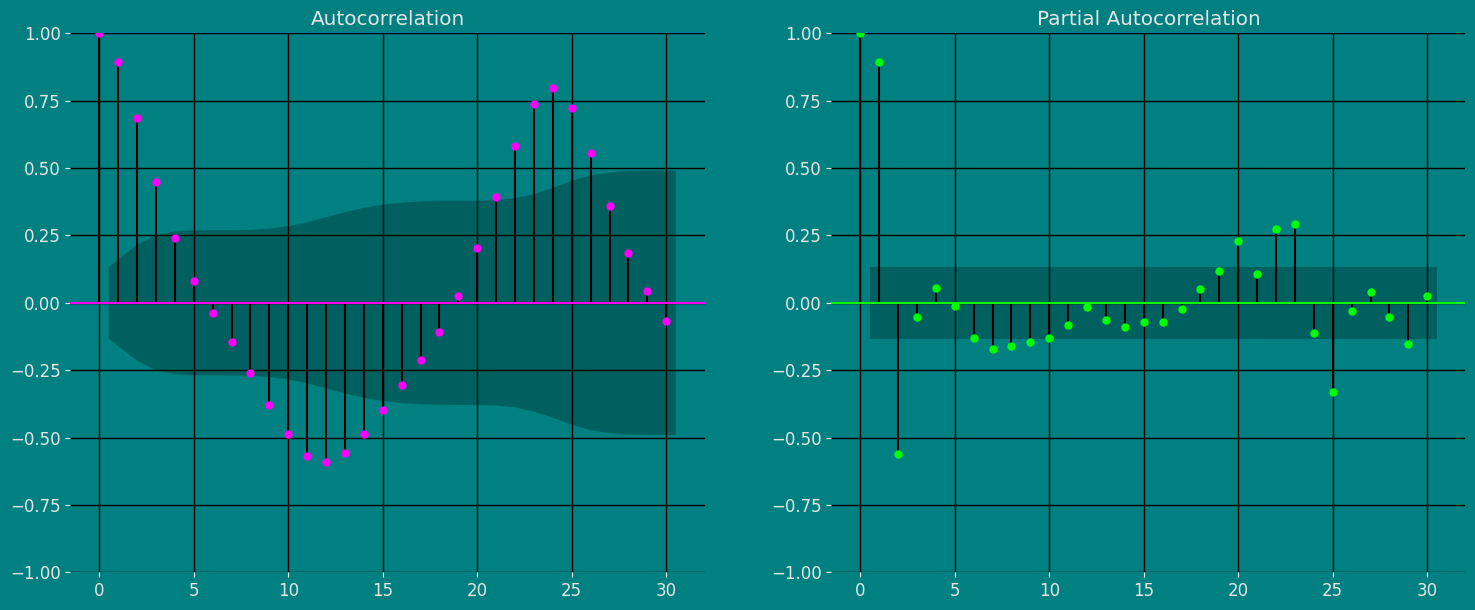
Autocorrelation (ACF) and partial autocorrelation (PACF) plots are statistical tools used to analyze time series. ACF charts show the correlation between the values of a time series and their lagged values, while PACF charts show the correlation between the values of a time series and their lagged values, after the effect of previous lagged values has been removed. ACF and PACF charts can be used to identify the structure of a time series, which can be helpful in choosing a suitable model for the time series. For example, if the ACF chart shows a repeating peak and valley pattern, this indicates that the time series is stationary, meaning that it has the same statistical properties over time. If the PACF chart shows a pattern of rapidly decreasing spikes, this indicates that the time series is invertible, meaning it can be reversed to get a stationary time series. The importance of the ACF and PACF charts is that they can help analysts better understand the structure of a time series. This understanding can be helpful in choosing a suitable model for the time series, which can improve the ability to predict future values of the time series. To analyze ACF and PACF charts:- Look for patterns in charts. Common patterns include repeating peaks and valleys, sawtooth patterns, and plateau patterns.
- Compare ACF and PACF charts. The PACF chart generally has fewer spikes than the ACF chart.
- Consider the length of the time series. ACF and PACF charts for longer time series will have more spikes.
- Use a confidence interval. The ACF and PACF plots also show confidence intervals for the autocorrelation values. If an autocorrelation value is outside the confidence interval, it is likely to be significant.
Decomposition of the time series
How to decompose a time series and why? In time series analysis to forecast new values, it is very important to know past data. More formally, we can say that it is very important to know the patterns that values follow over time. There can be many reasons that cause our forecast values to fall in the wrong direction. Basically, a time series consists of four components. The variation of those components causes the change in the pattern of the time series. These components are:- Level: This is the primary value that averages over time.
- Trend: The trend is the value that causes increasing or decreasing patterns in a time series.
- Seasonality: This is a cyclical event that occurs in a time series for a short time and causes short-term increasing or decreasing patterns in a time series.
- Residual/Noise: These are the random variations in the time series.
Additive time series
If the components of the time series are added to make the time series. Then the time series is called the additive time series. By visualization, we can say that the time series is additive if the increasing or decreasing pattern of the time series is similar throughout the series. The mathematical function of any additive time series can be represented by:Multiplicative time series
If the components of the time series are multiplicative together, then the time series is called a multiplicative time series. For visualization, if the time series is having exponential growth or decline with time, then the time series can be considered as the multiplicative time series. The mathematical function of the multiplicative time series can be represented as.Split the data into training and testing
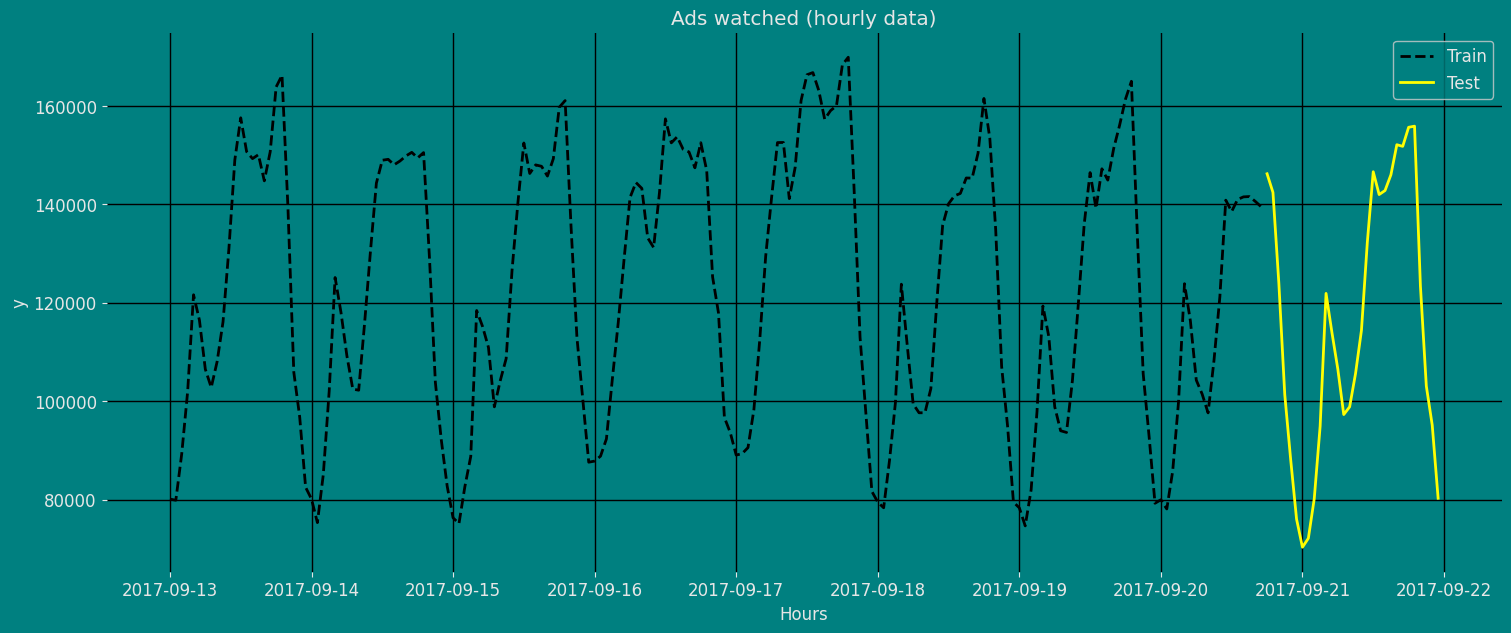
Let’s divide our data into sets 1. Data to train ourMSTL Model. 2.
Data to test our model
For the test data we will use the last 30 Hours to test and evaluate the
performance of our model.
Implementation of MSTL Method with StatsForecast
Load libraries
Instantiating Model
Import and instantiate the models. Setting the argument is sometimes tricky. This article on Seasonal periods by the master, Rob Hyndmann, can be useful forseason_length.
First, we must define the model parameters. As mentioned before, the
Candy production load presents seasonalities every 24 hours (Hourly) and
every 24 * 7 (Daily) hours. Therefore, we will use [24, 24 * 7] for
season length. The trend component will be forecasted with an
AutoARIMA model. (You can also try with: AutoTheta, AutoCES, and
AutoETS)
-
freq:a string indicating the frequency of the data. (See pandas’ available frequencies.) -
n_jobs:n_jobs: int, number of jobs used in the parallel processing, use -1 for all cores. -
fallback_model:a model to be used if a model fails.
Fit Model
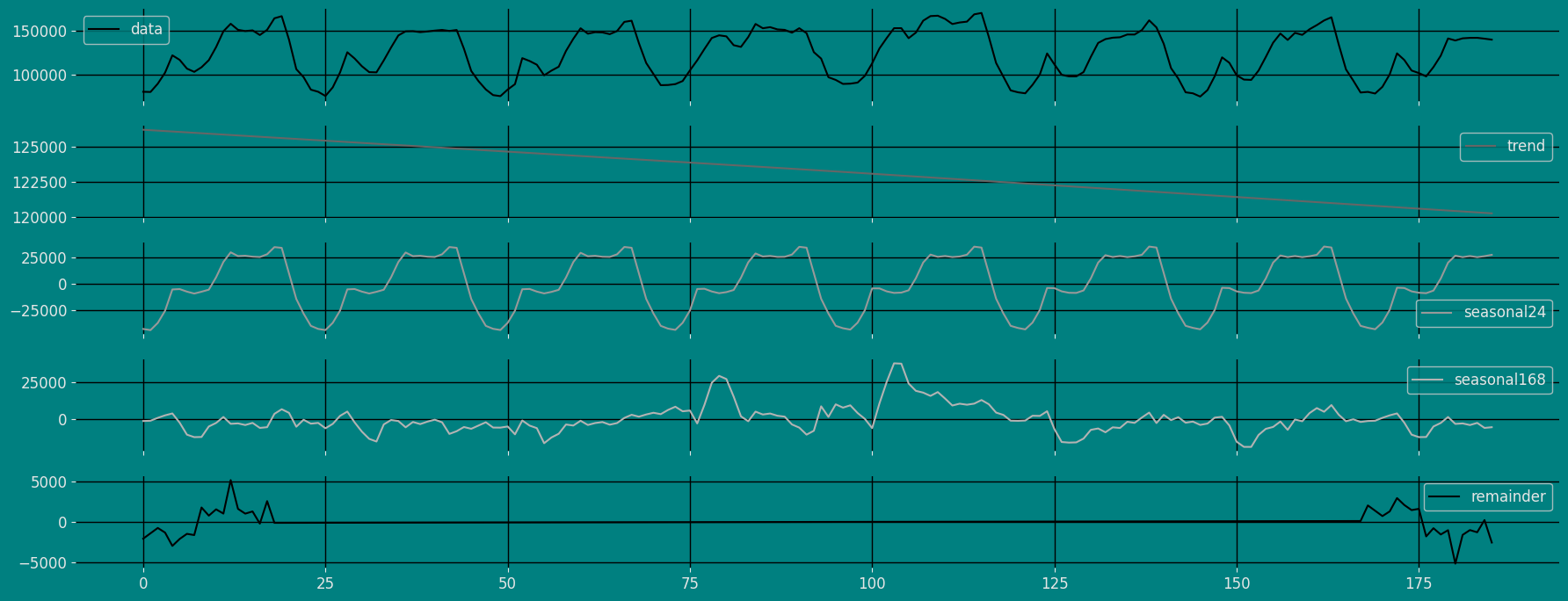
MSTL Model. We can observe it with the
following instruction:
| data | trend | seasonal24 | seasonal168 | remainder | |
|---|---|---|---|---|---|
| 0 | 80115.0 | 126222.558267 | -42511.086107 | -1524.379074 | -2072.093085 |
| 1 | 79885.0 | 126191.340644 | -43585.928105 | -1315.292640 | -1405.119899 |
| 2 | 89325.0 | 126160.117727 | -36756.458517 | 659.187427 | -737.846637 |
| … | … | … | … | … | … |
| 183 | 141590.0 | 120314.325647 | 25363.015190 | -2808.715638 | -1278.625199 |
| 184 | 140610.0 | 120280.850692 | 26306.688690 | -6221.712712 | 244.173330 |
| 185 | 139515.0 | 120247.361703 | 27571.777796 | -5745.053631 | -2559.085868 |
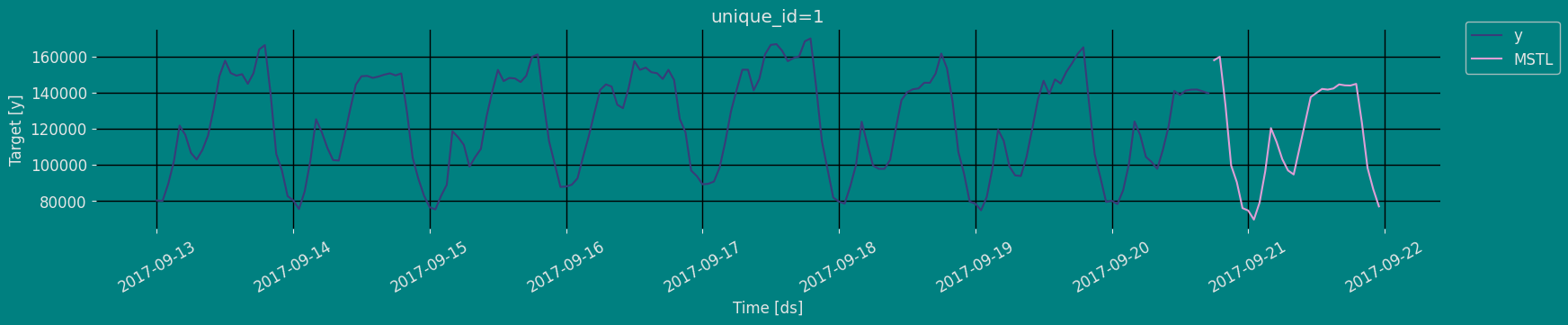
Forecast Method
If you want to gain speed in productive settings where you have multiple series or models we recommend using theStatsForecast.forecast method
instead of .fit and .predict.
The main difference is that the .forecast doest not store the fitted
values and is highly scalable in distributed environments.
The forecast method takes two arguments: forecasts next h (horizon)
and level.
-
h (int):represents the forecast h steps into the future. In this case, 30 hours ahead. -
level (list of floats):this optional parameter is used for probabilistic forecasting. Set the level (or confidence percentile) of your prediction interval. For example,level=[90]means that the model expects the real value to be inside that interval 90% of the times.
ARIMA and Theta)
| unique_id | ds | MSTL | |
|---|---|---|---|
| 0 | 1 | 2017-09-20 18:00:00 | 157848.500000 |
| 1 | 1 | 2017-09-20 19:00:00 | 159790.328125 |
| 2 | 1 | 2017-09-20 20:00:00 | 133002.281250 |
| … | … | … | … |
| 27 | 1 | 2017-09-21 21:00:00 | 98109.875000 |
| 28 | 1 | 2017-09-21 22:00:00 | 86342.015625 |
| 29 | 1 | 2017-09-21 23:00:00 | 76815.976562 |
| unique_id | ds | y | MSTL | |
|---|---|---|---|---|
| 0 | 1 | 2017-09-13 00:00:00 | 80115.0 | 79990.851562 |
| 1 | 1 | 2017-09-13 01:00:00 | 79885.0 | 79329.132812 |
| 2 | 1 | 2017-09-13 02:00:00 | 89325.0 | 88401.179688 |
| 3 | 1 | 2017-09-13 03:00:00 | 101930.0 | 102109.929688 |
| 4 | 1 | 2017-09-13 04:00:00 | 121630.0 | 123543.671875 |
| unique_id | ds | MSTL | MSTL-lo-95 | MSTL-hi-95 | |
|---|---|---|---|---|---|
| 0 | 1 | 2017-09-20 18:00:00 | 157848.500000 | 157796.406250 | 157900.593750 |
| 1 | 1 | 2017-09-20 19:00:00 | 159790.328125 | 159714.218750 | 159866.437500 |
| 2 | 1 | 2017-09-20 20:00:00 | 133002.281250 | 132893.937500 | 133110.609375 |
| … | … | … | … | … | … |
| 27 | 1 | 2017-09-21 21:00:00 | 98109.875000 | 95957.031250 | 100262.726562 |
| 28 | 1 | 2017-09-21 22:00:00 | 86342.015625 | 85410.578125 | 87273.460938 |
| 29 | 1 | 2017-09-21 23:00:00 | 76815.976562 | 73476.195312 | 80155.757812 |
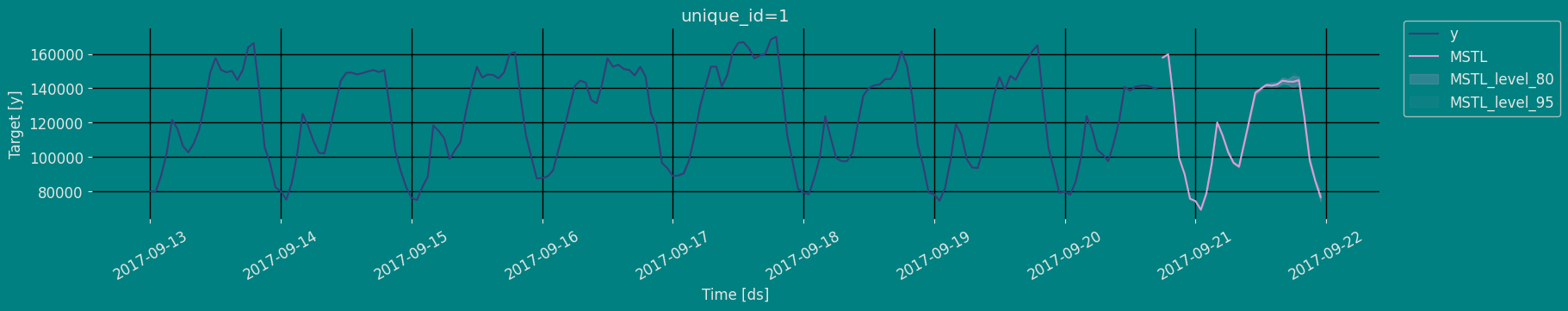
Predict method with confidence interval
To generate forecasts use the predict method. The predict method takes two arguments: forecasts the nexth (for
horizon) and level.
-
h (int):represents the forecast h steps into the future. In this case, 30 hours ahead. -
level (list of floats):this optional parameter is used for probabilistic forecasting. Set the level (or confidence percentile) of your prediction interval. For example,level=[95]means that the model expects the real value to be inside that interval 95% of the times.
| unique_id | ds | MSTL | |
|---|---|---|---|
| 0 | 1 | 2017-09-20 18:00:00 | 157848.500000 |
| 1 | 1 | 2017-09-20 19:00:00 | 159790.328125 |
| 2 | 1 | 2017-09-20 20:00:00 | 133002.281250 |
| … | … | … | … |
| 27 | 1 | 2017-09-21 21:00:00 | 98109.875000 |
| 28 | 1 | 2017-09-21 22:00:00 | 86342.015625 |
| 29 | 1 | 2017-09-21 23:00:00 | 76815.976562 |
| unique_id | ds | MSTL | MSTL-lo-95 | MSTL-lo-80 | MSTL-hi-80 | MSTL-hi-95 | |
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2017-09-20 18:00:00 | 157848.500000 | 157796.406250 | 157798.484375 | 157898.531250 | 157900.593750 |
| 1 | 1 | 2017-09-20 19:00:00 | 159790.328125 | 159714.218750 | 159716.187500 | 159864.468750 | 159866.437500 |
| 2 | 1 | 2017-09-20 20:00:00 | 133002.281250 | 132893.937500 | 132894.515625 | 133110.031250 | 133110.609375 |
| … | … | … | … | … | … | … | … |
| 27 | 1 | 2017-09-21 21:00:00 | 98109.875000 | 95957.031250 | 96493.921875 | 99725.828125 | 100262.726562 |
| 28 | 1 | 2017-09-21 22:00:00 | 86342.015625 | 85410.578125 | 85411.835938 | 87272.195312 | 87273.460938 |
| 29 | 1 | 2017-09-21 23:00:00 | 76815.976562 | 73476.195312 | 74494.546875 | 79137.406250 | 80155.757812 |
Cross-validation
In previous steps, we’ve taken our historical data to predict the future. However, to asses its accuracy we would also like to know how the model would have performed in the past. To assess the accuracy and robustness of your models on your data perform Cross-Validation. With time series data, Cross Validation is done by defining a sliding window across the historical data and predicting the period following it. This form of cross-validation allows us to arrive at a better estimation of our model’s predictive abilities across a wider range of temporal instances while also keeping the data in the training set contiguous as is required by our models. The following graph depicts such a Cross Validation Strategy: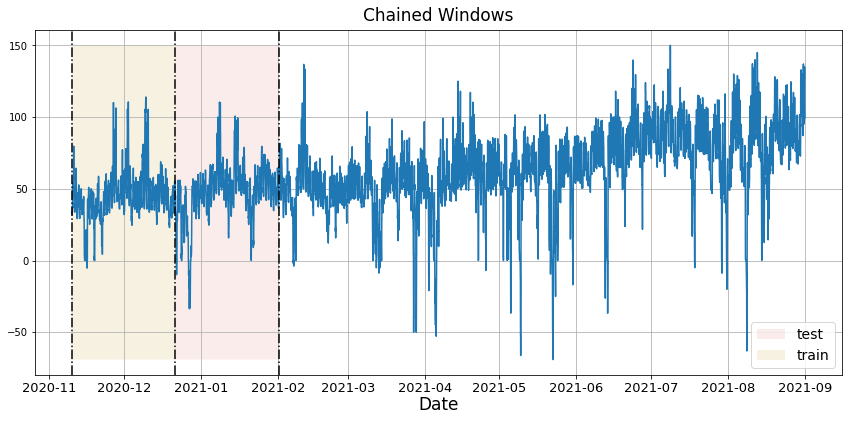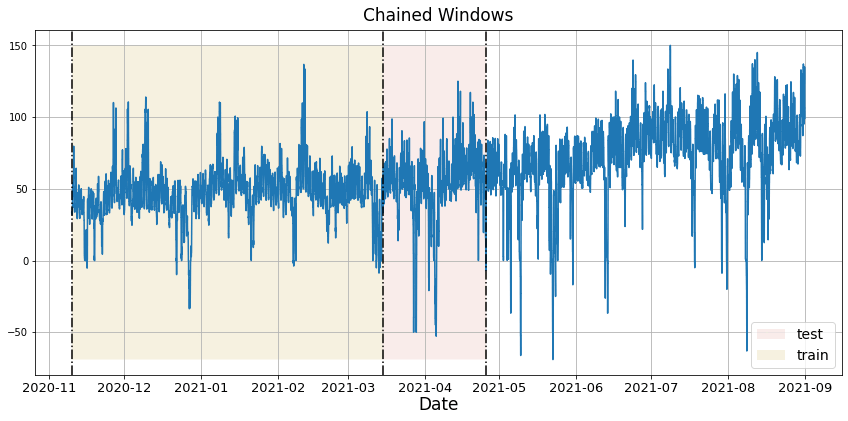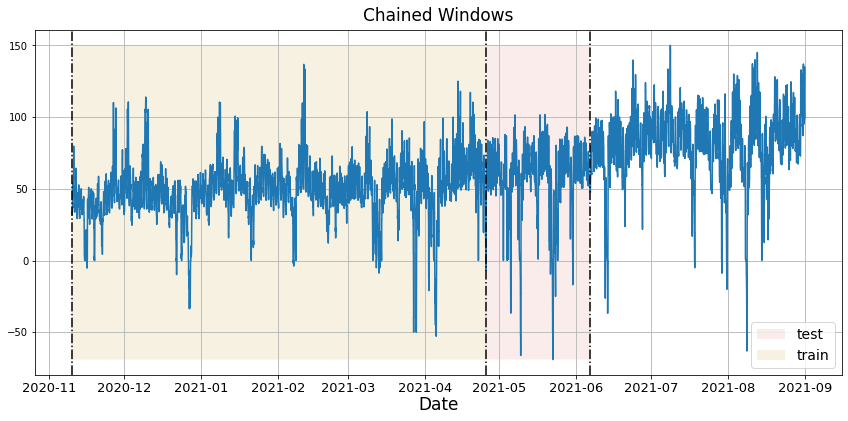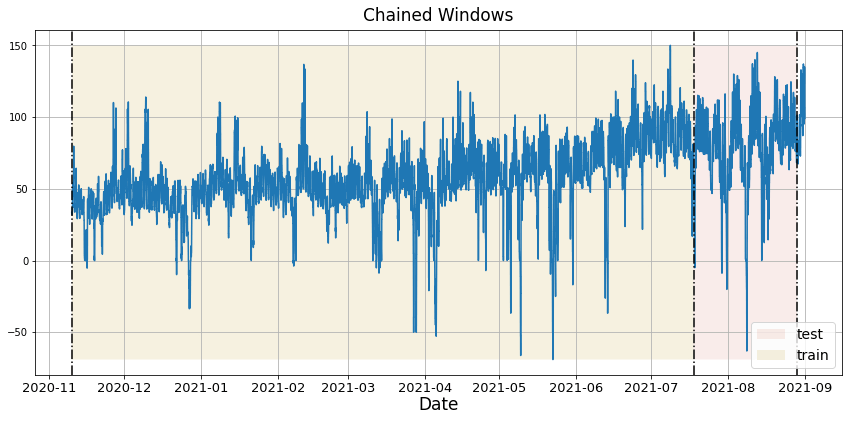
Perform time series cross-validation
Cross-validation of time series models is considered a best practice but most implementations are very slow. The statsforecast library implements cross-validation as a distributed operation, making the process less time-consuming to perform. If you have big datasets you can also perform Cross Validation in a distributed cluster using Ray, Dask or Spark. In this case, we want to evaluate the performance of each model for the last 5 months(n_windows=), forecasting every second months
(step_size=50). Depending on your computer, this step should take
around 1 min.
The cross_validation method from the StatsForecast class takes the
following arguments.
-
df:training data frame -
h (int):represents h steps into the future that are being forecasted. In this case, 500 hours ahead. -
step_size (int):step size between each window. In other words: how often do you want to run the forecasting processes. -
n_windows(int):number of windows used for cross validation. In other words: what number of forecasting processes in the past do you want to evaluate.
unique_id:series identifierds:datestamp or temporal indexcutoff:the last datestamp or temporal index for then_windows.y:true valuemodel:columns with the model’s name and fitted value.
| unique_id | ds | cutoff | y | MSTL | |
|---|---|---|---|---|---|
| 0 | 1 | 2017-09-15 18:00:00 | 2017-09-15 17:00:00 | 159725.0 | 158384.250000 |
| 1 | 1 | 2017-09-15 19:00:00 | 2017-09-15 17:00:00 | 161085.0 | 162015.171875 |
| 2 | 1 | 2017-09-15 20:00:00 | 2017-09-15 17:00:00 | 135520.0 | 138495.093750 |
| … | … | … | … | … | … |
| 147 | 1 | 2017-09-21 21:00:00 | 2017-09-20 17:00:00 | 103080.0 | 98109.875000 |
| 148 | 1 | 2017-09-21 22:00:00 | 2017-09-20 17:00:00 | 95155.0 | 86342.015625 |
| 149 | 1 | 2017-09-21 23:00:00 | 2017-09-20 17:00:00 | 80285.0 | 76815.976562 |
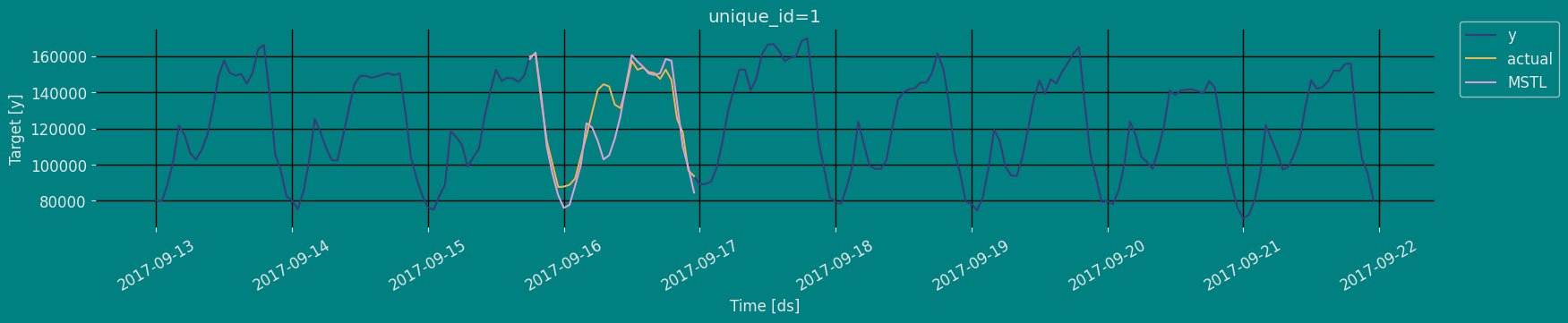
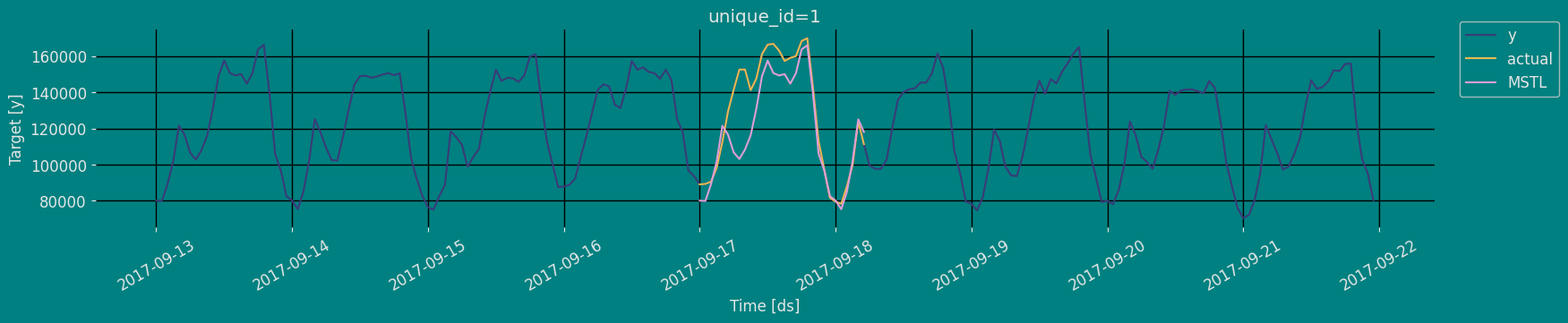
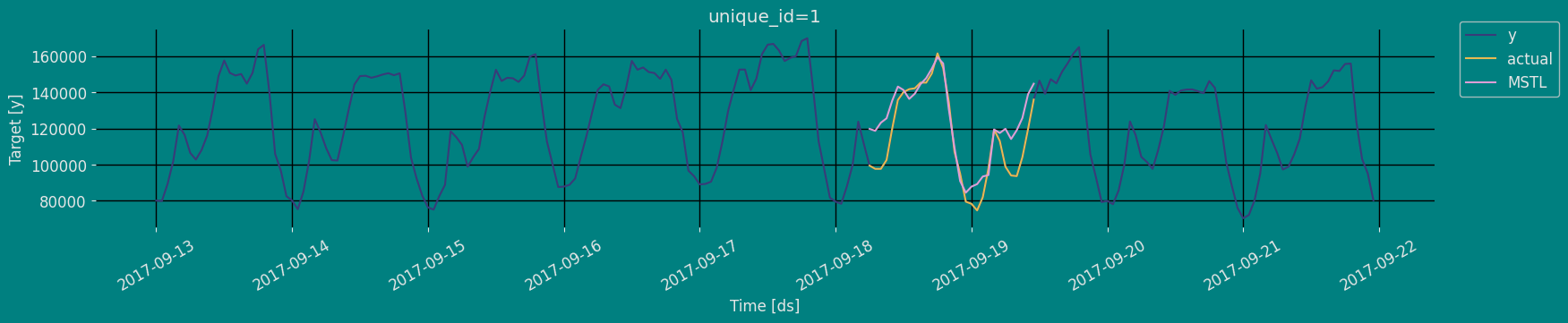
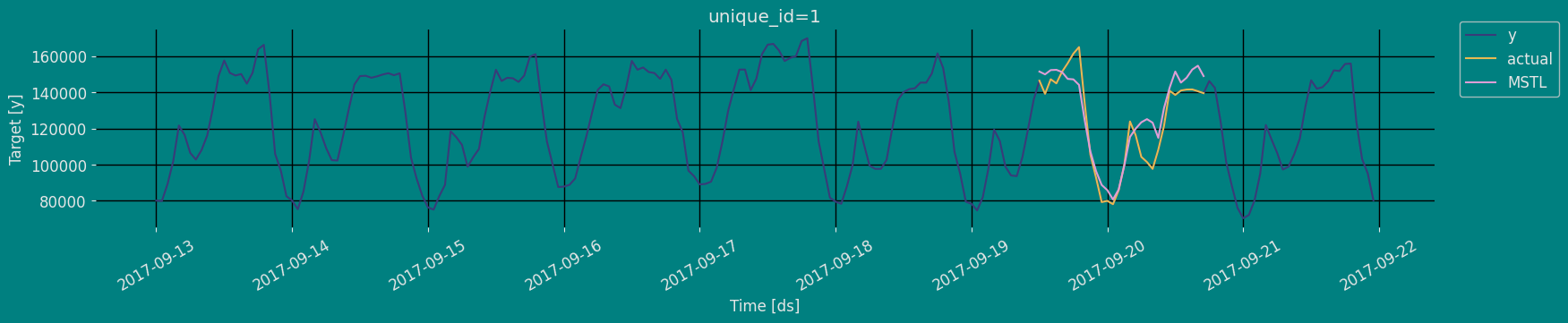
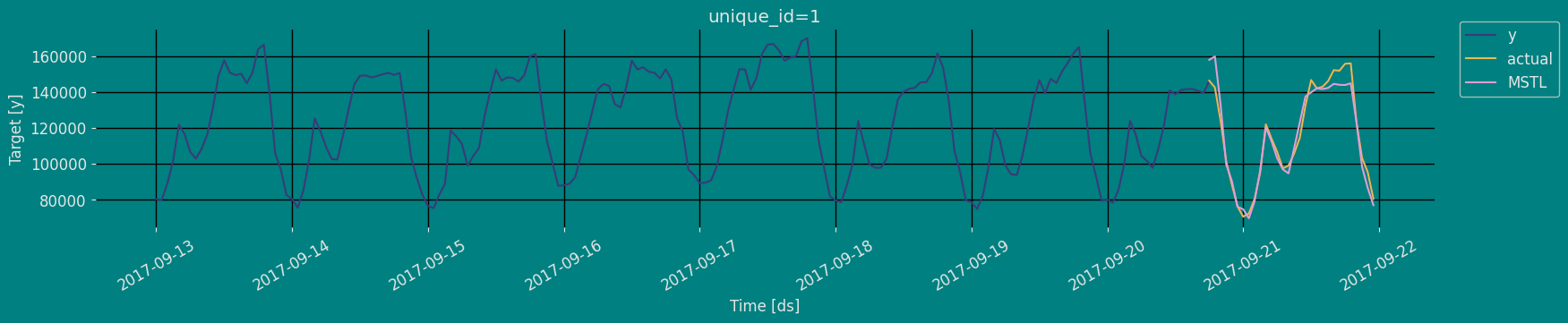
Model Evaluation
Now we are going to evaluate our model with the results of the predictions, we will use different types of metrics MAE, MAPE, MASE, RMSE, SMAPE to evaluate the accuracy.| unique_id | metric | MSTL | |
|---|---|---|---|
| 0 | 1 | mae | 4932.395052 |
| 1 | 1 | mape | 0.040514 |
| 2 | 1 | mase | 0.609407 |
| 3 | 1 | rmse | 6495.207028 |
| 4 | 1 | smape | 0.020267 |
References
- Changquan Huang • Alla Petukhina. Springer series (2022). Applied Time Series Analysis and Forecasting with Python.
- Ivan Svetunkov. Forecasting and Analytics with the Augmented Dynamic Adaptive Model (ADAM)
- James D. Hamilton. Time Series Analysis Princeton University Press, Princeton, New Jersey, 1st Edition, 1994.
- Nixtla MultipleSeasonalTrend API
- Pandas available frequencies.
- Rob J. Hyndman and George Athanasopoulos (2018). “Forecasting Principles and Practice (3rd ed)”.
- Seasonal periods- Rob J Hyndman.