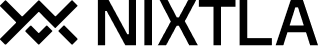Detailed description of all the functionalities that MLForecast provides.
Data setup
For this example we’ll use a subset of the M4 hourly dataset. You can find the a notebook with the full dataset here.| unique_id | ds | y | |
|---|---|---|---|
| 0 | H196 | 1 | 11.8 |
| 1 | H196 | 2 | 11.4 |
| 2 | H196 | 3 | 11.1 |
| 3 | H196 | 4 | 10.8 |
| 4 | H196 | 5 | 10.6 |
| … | … | … | … |
| 4027 | H413 | 1004 | 99.0 |
| 4028 | H413 | 1005 | 88.0 |
| 4029 | H413 | 1006 | 47.0 |
| 4030 | H413 | 1007 | 41.0 |
| 4031 | H413 | 1008 | 34.0 |
EDA
We’ll take a look at our series to get ideas for transformations and features.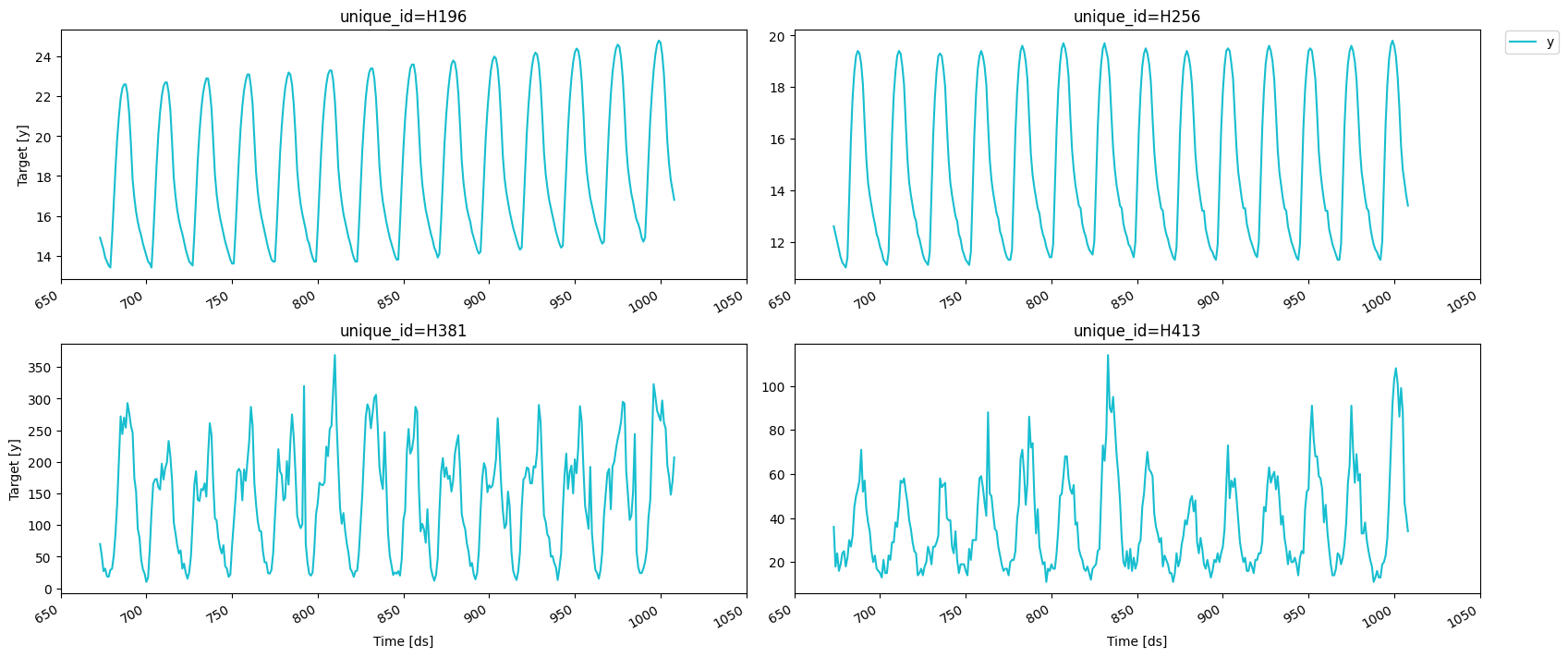
MLForecast.preprocess method to explore different
transformations. It looks like these series have a strong seasonality on
the hour of the day, so we can subtract the value from the same hour in
the previous day to remove it. This can be done with the
mlforecast.target_transforms.Differences transformer, which we pass
through target_transforms.
| unique_id | ds | y | |
|---|---|---|---|
| 24 | H196 | 25 | 0.3 |
| 25 | H196 | 26 | 0.3 |
| 26 | H196 | 27 | 0.1 |
| 27 | H196 | 28 | 0.2 |
| 28 | H196 | 29 | 0.2 |
| … | … | … | … |
| 4027 | H413 | 1004 | 39.0 |
| 4028 | H413 | 1005 | 55.0 |
| 4029 | H413 | 1006 | 14.0 |
| 4030 | H413 | 1007 | 3.0 |
| 4031 | H413 | 1008 | 4.0 |
Adding features
Lags
Looks like the seasonality is gone, we can now try adding some lag features.| unique_id | ds | y | lag1 | lag24 | |
|---|---|---|---|---|---|
| 48 | H196 | 49 | 0.1 | 0.1 | 0.3 |
| 49 | H196 | 50 | 0.1 | 0.1 | 0.3 |
| 50 | H196 | 51 | 0.2 | 0.1 | 0.1 |
| 51 | H196 | 52 | 0.1 | 0.2 | 0.2 |
| 52 | H196 | 53 | 0.1 | 0.1 | 0.2 |
| … | … | … | … | … | … |
| 4027 | H413 | 1004 | 39.0 | 29.0 | 1.0 |
| 4028 | H413 | 1005 | 55.0 | 39.0 | -25.0 |
| 4029 | H413 | 1006 | 14.0 | 55.0 | -20.0 |
| 4030 | H413 | 1007 | 3.0 | 14.0 | 0.0 |
| 4031 | H413 | 1008 | 4.0 | 3.0 | -16.0 |
Lag transforms
Lag transforms are defined as a dictionary where the keys are the lags and the values are the transformations that we want to apply to that lag. The lag transformations can be either objects from themlforecast.lag_transforms module or numba
jitted functions (so that computing the features doesn’t become a
bottleneck and we can bypass the GIL when using multithreading), we have
some implemented in the window-ops
package but you can also
implement your own.
| unique_id | ds | y | expanding_mean_lag1 | rolling_mean_lag24_window_size48 | rolling_mean_48_lag24 | |
|---|---|---|---|---|---|---|
| 95 | H196 | 96 | 0.1 | 0.174648 | 0.150000 | 0.150000 |
| 96 | H196 | 97 | 0.3 | 0.173611 | 0.145833 | 0.145833 |
| 97 | H196 | 98 | 0.3 | 0.175342 | 0.141667 | 0.141667 |
| 98 | H196 | 99 | 0.3 | 0.177027 | 0.141667 | 0.141667 |
| 99 | H196 | 100 | 0.3 | 0.178667 | 0.141667 | 0.141667 |
| … | … | … | … | … | … | … |
| 4027 | H413 | 1004 | 39.0 | 0.242084 | 3.437500 | 3.437500 |
| 4028 | H413 | 1005 | 55.0 | 0.281633 | 2.708333 | 2.708333 |
| 4029 | H413 | 1006 | 14.0 | 0.337411 | 2.125000 | 2.125000 |
| 4030 | H413 | 1007 | 3.0 | 0.351324 | 1.770833 | 1.770833 |
| 4031 | H413 | 1008 | 4.0 | 0.354018 | 1.208333 | 1.208333 |
Date features
If your time column is made of timestamps then it might make sense to extract features like week, dayofweek, quarter, etc. You can do that by passing a list of strings with pandas time/date components. You can also pass functions that will take the time column as input, as we’ll show here.| unique_id | ds | y | hour_index | |
|---|---|---|---|---|
| 24 | H196 | 25 | 0.3 | 1 |
| 25 | H196 | 26 | 0.3 | 2 |
| 26 | H196 | 27 | 0.1 | 3 |
| 27 | H196 | 28 | 0.2 | 4 |
| 28 | H196 | 29 | 0.2 | 5 |
| … | … | … | … | … |
| 4027 | H413 | 1004 | 39.0 | 20 |
| 4028 | H413 | 1005 | 55.0 | 21 |
| 4029 | H413 | 1006 | 14.0 | 22 |
| 4030 | H413 | 1007 | 3.0 | 23 |
| 4031 | H413 | 1008 | 4.0 | 0 |
Target transformations
If you want to do some transformation to your target before computing the features and then re-apply it after predicting you can use thetarget_transforms argument, which takes a list of transformations. You
can find the implemented ones in mlforecast.target_transforms or you
can implement your own as described in the target transformations
guide.
| unique_id | ds | y | lag1 | |
|---|---|---|---|---|
| 1 | H196 | 2 | -1.493026 | -1.383286 |
| 2 | H196 | 3 | -1.575331 | -1.493026 |
| 3 | H196 | 4 | -1.657635 | -1.575331 |
| 4 | H196 | 5 | -1.712505 | -1.657635 |
| 5 | H196 | 6 | -1.794810 | -1.712505 |
| … | … | … | … | … |
| 4027 | H413 | 1004 | 3.062766 | 2.425012 |
| 4028 | H413 | 1005 | 2.523128 | 3.062766 |
| 4029 | H413 | 1006 | 0.511751 | 2.523128 |
| 4030 | H413 | 1007 | 0.217403 | 0.511751 |
| 4031 | H413 | 1008 | -0.126003 | 0.217403 |
| unique_id | ds | Naive | |
|---|---|---|---|
| 0 | H196 | 1009 | 16.8 |
| 1 | H256 | 1009 | 13.4 |
| 2 | H381 | 1009 | 207.0 |
| 3 | H413 | 1009 | 34.0 |
| unique_id | ds | y | |
|---|---|---|---|
| 1007 | H196 | 1008 | 16.8 |
| 2015 | H256 | 1008 | 13.4 |
| 3023 | H381 | 1008 | 207.0 |
| 4031 | H413 | 1008 | 34.0 |
Training
Once you’ve decided the features, transformations and models that you want to use you can use theMLForecast.fit method instead, which will
do the preprocessing and then train the models. The models can be
specified as a list (which will name them by using their class name and
an index if there are repeated classes) or as a dictionary where the
keys are the names you want to give to the models, i.e. the name of the
column that will hold their predictions, and the values are the models
themselves.
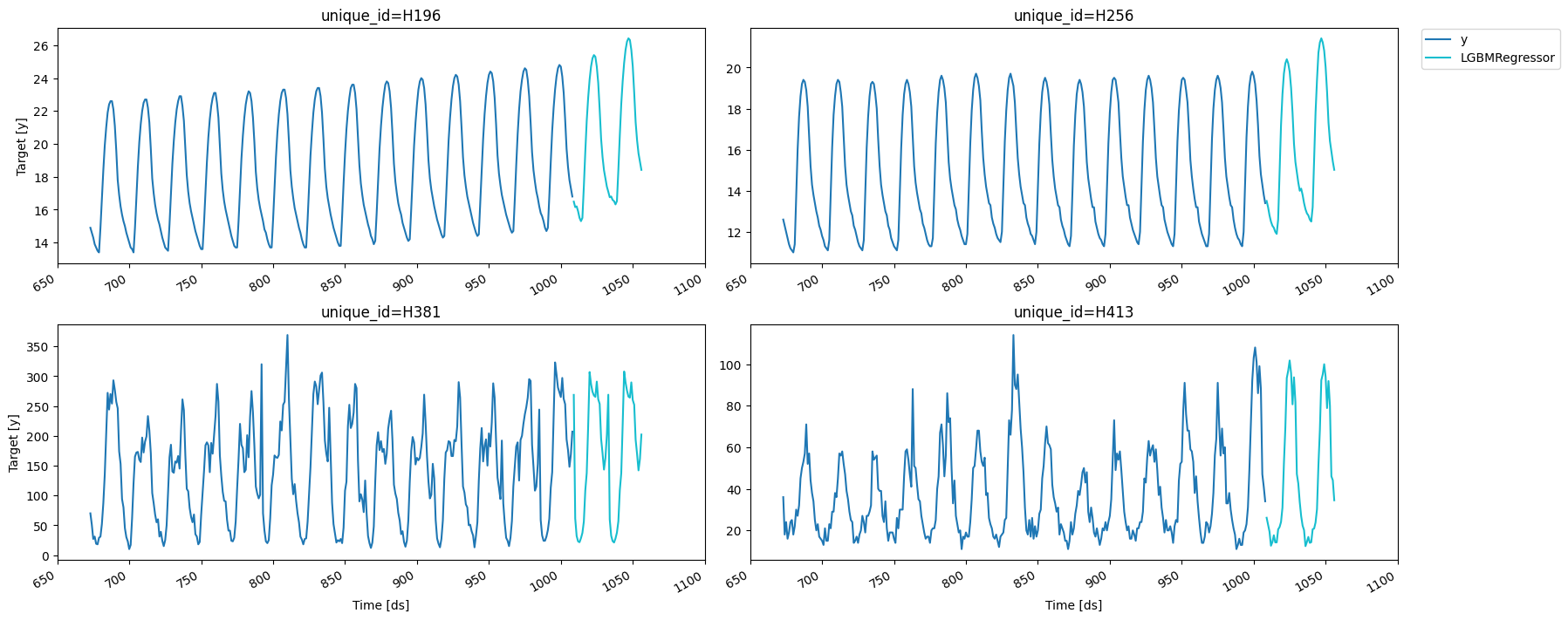
Forecasting
| unique_id | ds | avg | q75 | q25 | |
|---|---|---|---|---|---|
| 0 | H196 | 1009 | 16.295257 | 16.357148 | 16.315731 |
| 1 | H196 | 1010 | 15.910282 | 16.007322 | 15.862261 |
| 2 | H196 | 1011 | 15.728367 | 15.780183 | 15.658180 |
| 3 | H196 | 1012 | 15.468414 | 15.513598 | 15.399717 |
| 4 | H196 | 1013 | 15.081279 | 15.133848 | 15.007694 |
| … | … | … | … | … | … |
| 187 | H413 | 1052 | 100.450617 | 124.211150 | 47.025017 |
| 188 | H413 | 1053 | 88.426800 | 108.303409 | 44.715380 |
| 189 | H413 | 1054 | 59.675737 | 81.859964 | 19.239462 |
| 190 | H413 | 1055 | 57.580356 | 72.703301 | 21.486674 |
| 191 | H413 | 1056 | 42.669879 | 46.018271 | 24.392357 |
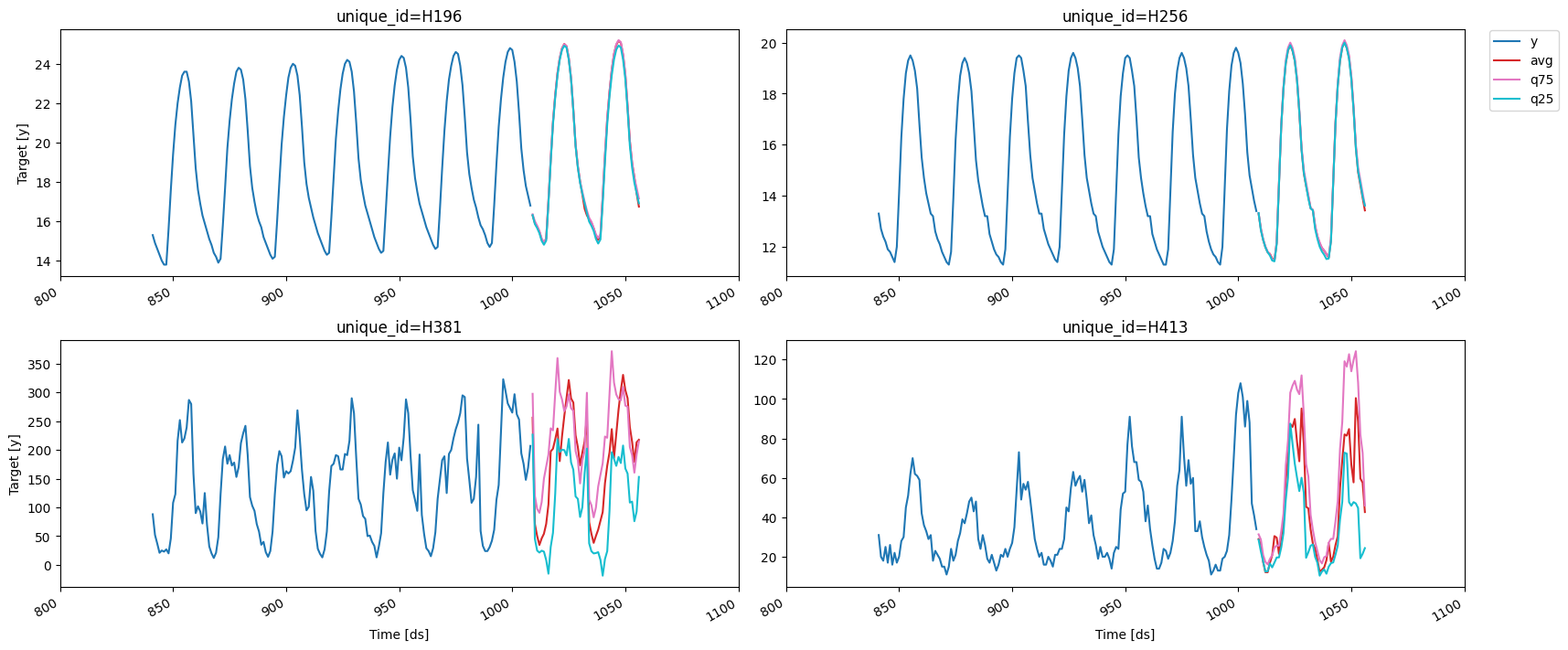
Saving and loading
The MLForecast class has theMLForecast.save and MLForecast.load to
store and then load the forecast object.
Updating series’ values
After you’ve trained a forecast object you can save and load it with the previous methods. If by the time you want to use it you already know the following values of the target you can use theMLForecast.update
method to incorporate these, which will allow you to use these new
values when computing predictions.
- If no new values are provided for a series that’s currently stored, only the previous ones are kept.
- If new series are included they are added to the existing ones.
| unique_id | ds | Naive | |
|---|---|---|---|
| 0 | H196 | 1009 | 16.8 |
| 1 | H256 | 1009 | 13.4 |
| 2 | H381 | 1009 | 207.0 |
| 3 | H413 | 1009 | 34.0 |
| unique_id | ds | Naive | |
|---|---|---|---|
| 0 | H196 | 1010 | 17.0 |
| 1 | H256 | 1010 | 14.0 |
| 2 | H381 | 1009 | 207.0 |
| 3 | H413 | 1009 | 34.0 |
Estimating model performance
Cross validation
In order to get an estimate of how well our model will be when predicting future data we can perform cross validation, which consists of training a few models independently on different subsets of the data, using them to predict a validation set and measuring their performance. Since our data depends on time, we make our splits by removing the last portions of the series and using them as validation sets. This process is implemented inMLForecast.cross_validation.
| unique_id | ds | cutoff | y | LGBMRegressor | |
|---|---|---|---|---|---|
| 0 | H196 | 817 | 816 | 15.3 | 15.383165 |
| 1 | H196 | 818 | 816 | 14.9 | 14.923219 |
| 2 | H196 | 819 | 816 | 14.6 | 14.667834 |
| 3 | H196 | 820 | 816 | 14.2 | 14.275964 |
| 4 | H196 | 821 | 816 | 13.9 | 13.973491 |
| … | … | … | … | … | … |
| 763 | H413 | 1004 | 960 | 99.0 | 65.644823 |
| 764 | H413 | 1005 | 960 | 88.0 | 71.717097 |
| 765 | H413 | 1006 | 960 | 47.0 | 76.704377 |
| 766 | H413 | 1007 | 960 | 41.0 | 53.446638 |
| 767 | H413 | 1008 | 960 | 34.0 | 54.902634 |
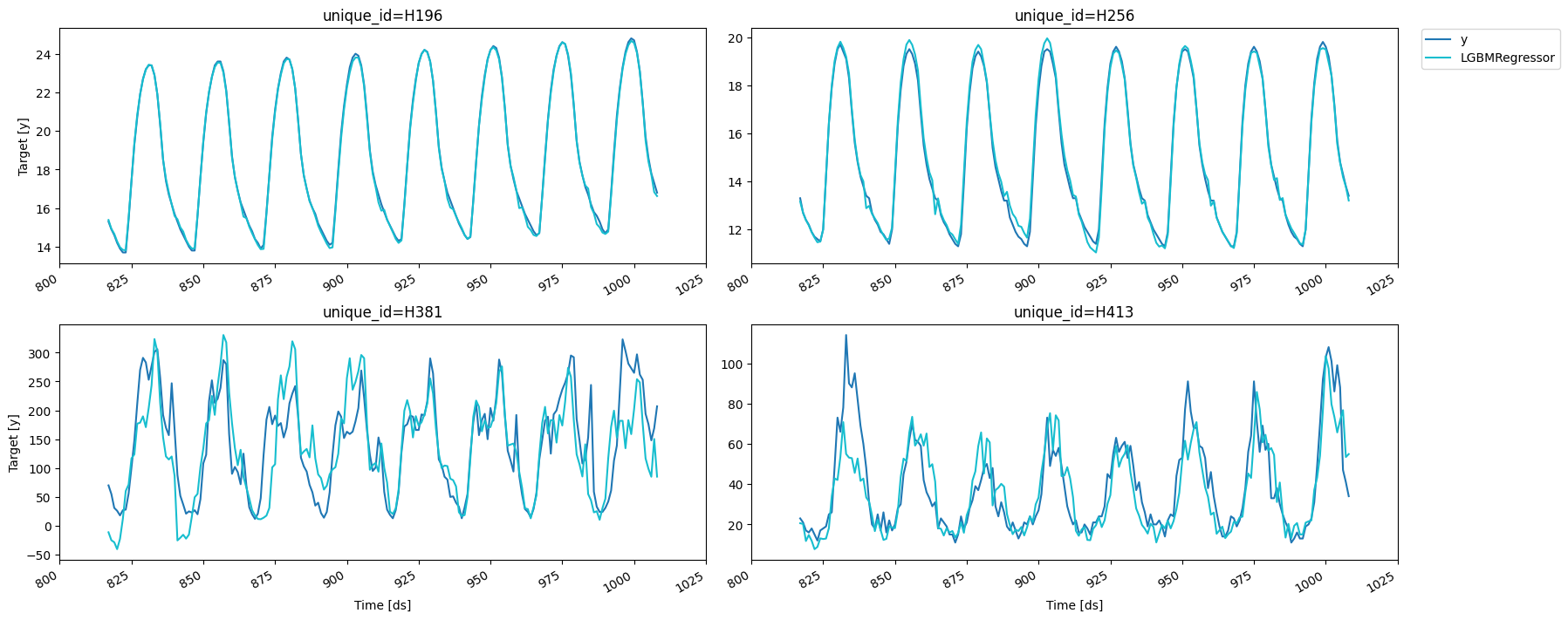
| LGBMRegressor | |
|---|---|
| cutoff | |
| 816 | 29.418172 |
| 864 | 34.257598 |
| 912 | 13.145763 |
| 960 | 35.066261 |
LightGBMCV
In the same spirit of estimating our model’s performance,LightGBMCV
allows us to train a few
LightGBM models on different
partitions of the data. The main differences with
MLForecast.cross_validation are:
- It can only train LightGBM models.
- It trains all models simultaneously and gives us per-iteration averages of the errors across the complete forecasting window, which allows us to find the best iteration.
eval_every argument) and performs early stopping (which can be
configured with early_stopping_evals and early_stopping_pct). If you
set compute_cv_preds=True the out-of-fold predictions are computed
using the best iteration found and are saved in the cv_preds_
attribute.
| unique_id | ds | y | Booster | window | |
|---|---|---|---|---|---|
| 0 | H196 | 817 | 15.3 | 15.473182 | 0 |
| 1 | H196 | 818 | 14.9 | 15.038571 | 0 |
| 2 | H196 | 819 | 14.6 | 14.849409 | 0 |
| 3 | H196 | 820 | 14.2 | 14.448379 | 0 |
| 4 | H196 | 821 | 13.9 | 14.148379 | 0 |
| … | … | … | … | … | … |
| 187 | H413 | 1004 | 99.0 | 61.425396 | 3 |
| 188 | H413 | 1005 | 88.0 | 62.886890 | 3 |
| 189 | H413 | 1006 | 47.0 | 57.886890 | 3 |
| 190 | H413 | 1007 | 41.0 | 38.849009 | 3 |
| 191 | H413 | 1008 | 34.0 | 44.720562 | 3 |
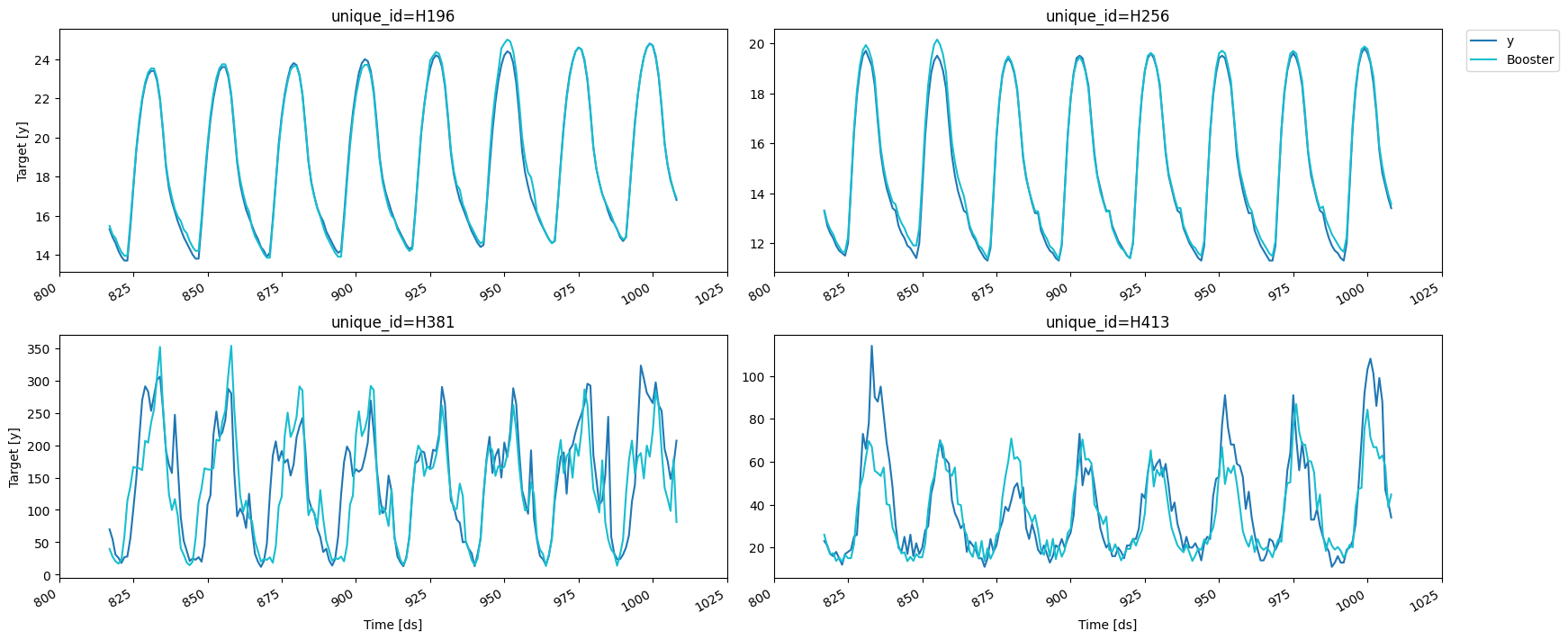
MLForecast object from the LightGBMCV one as
follows: